
GenOak
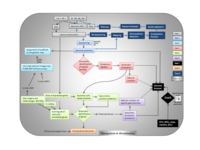
Schematic overview of the GENOAK project structure
Sequencing of the oak genome and identification of genes that matter for forest tree adaptation
Call: ANR "Blanc"
Trees are an important component of the global ecosystem. They dominate many regions of the world as natural, extensively or intensively managed plantations. The Fagaceae family comprises about 1,000 woody species distributed throughout the northern hemisphere. About half belong to the Quercus (oak) genus. These oaks have traditionally high cultural and societal values. As other forest tree species, they are also providing important environmental (carbon sequestration, water cycle, reservoir of biodiversity, soil protection...) and economic (carpentry, furniture, cabinet making, veneer, cask industry, fuelwood, hunting and fungus gathering) services. Finally these long-lived organisms are considered as relevant models to study both short and long term adaptation mechanisms to abiotic and biotic constraints associated with global change, as they grow under a wide range of environments (soil and climate). Due to their biological dominance, their commercial and ecological importance worldwide, an international network was launched recently to develop a coordinated strategy to sequence several Fagaceae genomes. These genomic resources will allow to discover which genes are involved in the adaptation of these organisms to their environments and to study their adaptive potential under current and future climate changes. In Europe, genomic resources have been developed for sessile and pedunculate oaks in the frame of the EVOLTREE network of excellence. In US, American and Chinese chestnuts (Castanea) were the main targets of a NSF funded project. Their genome is now being sequenced within the Forest Health Initiative (http://foresthealthinitiative.org/). In China, the efforts to sequence the Lithocarpus and Castanopsis genomes are also on the right track.
This project aims at supporting the participation of French Institutions involved in this international effort. It is a collaboration between four INRA research units and the GENOSCOPE: the French National Sequencing Center. Our objectives are three fold: 1/ to obtain a reference sequence for the pedunculate oak genome. Sequencing of a genome like oak (740Mb/C) is indeed becoming possible at a substantially reduced cost due to breakthroughs in sequencing technologies and advances in bioinformatics, 2/ annotate the genome and make the data and results available to the scientific community through interoperable databases, and 3/ identify genes that matter for forest tree adaptation through the analysis of adaptive divergence (incipient ecological speciation) at the transcriptome and genome-wide levels by respectively i) identifying differentially expressed genes between co-occuring species (sessile and pedunculate oaks) adapted to different edapho-climatic environments, and ii) analyzing the size, distribution and nature of genomic islands of divergence maintained in the face of gene flow of the same four sympatric species.
Duration: 01/10/2011 to 30/09/2015
Coordinator: Christophe Plomion
Partners
- UMR BIOGECO ( https://www4.bordeaux-aquitaine.inra.fr/biogeco/ ), Christophe Plomion
- GENOSCOPE ( http://www.genoscope.cns.fr/ ), Jean-Marc Aury
- URGI ( https://urgi.versailles.inra.fr/ ), Joëlle Amselem, Hadi Quesneville
- UMR GDEC, Clermont ( http://www1.clermont.inra.fr/umr1095/ ), JĂ©rome Salse
- UMR IAM Nancy ( http://mycor.nancy.inra.fr/ ), Francis Martin
Creation date: 28 Dec 2011
 eZ Publish
eZ PublishPublication supervisor: A-F. Adam-Blondon
Read Credits & General Terms of Use
Read How to cite








