
Ephesis
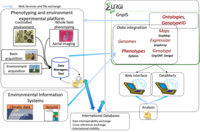
Ephesis, or Environment and Phenotype Information System, is an INRA URGI platform project. It has been initiated by the Departement Genetic et amelioration des Plantes (DGAP), which has ensured its long term stability by assigning this mission to a permanent engineer. It provides a national database storing Genotype by Environment experiment results, developed within the URGI information system, GnpIS.
The Ephesis project has produced a web application, Ephesis, which grant access to phenotype data stored in GnpIS. This application is available at the following address: https://urgi.versailles.inra.fr/ephesis/
Genotypes, Environment and Phenotypes Relationship studies at INRA
The investigation of the relationship between genotypes, environment and phenotypes is one of the main long term goals of the INRA. To attain this objective, researchers need a massive amount of data and tools to create, handle and finally analyze it. This observation has led the IGEC ( Interaction Genotype Environment and Cultural systems) scientific network to set up three axes of development :
- High throughput data generation through the use of new technologies, like aerial image processing or automated measurement tools. To fulfill this objective it is also critical to rationalize the basic acquisition of experimental data and cultural processes.
- Information System to store, filter and reorganise data. Multiple softwares are needed to answer this challenge : data management tool to ease data production and an integrative portal, Ephesis.
- Analysis tools (Statistical, mathematical, ...) to extract knowledge from the data.
Ephesis
Goals
The Ephesis information system is dedicated to the study of genotype by environment interactions (G*E). It increases the visibility of experiments conducted at the INRA, give value to experimental data acquired during specific temporal or spatial series to allow the analysis of genotypes to environmental constraints. The system eases data access and structuring, and the set up of meta analysis.This is reinforced by integrating experiments conducted by international partner alongside INRA data.
Integration of environmental is achieved in collaboration with INRA experts and especially with AgroClim and InfoSol. The integration in GnpIS provides a tight integration with ontologies and genetic resources traceability needed to set up cross experiment G*E data sets. Furthermore, germplasm identification allows to integrate Ephesis data with genotyping data sotred either in GnpIS or remotely and thus to build association genetic data sets.
Ephesis Functionalities
Current functionalities:
- Integration of Phenotypic experimentation Trial, with a tractability of phenotypes, environment, and relevant Trial information.
- Ontology driven genericity to manage the wide spectrum of existingTrials.
- Data quality ensured by link to expertized germplasm panels and collection.
- Querying, organisation, sorting and linking of genotypic, phenotypic and environmental data.
- Allow the most relevant level of traceability of raw and elaborate data.
- Support the construction and export of data set designed for analysis purpose.
Perspectives
Ephesis is still under very active development. Future improvement will focus on :
- Improved handling of environment data, including agricultural practice
- Improved data quality by integrating in GnpIS evolved ontologies and new germplasm collections.
- Integration within a national and international network of phenotype databases
- Adonis : basic data acquisition
- Local data management tool : raw data integration at the level of each experimental platforms (Ephesis partners, High Throughput phenotyping Phenome nodes, …)
- International networks (EBI Transplant, …)
Partners and collaboration.
The development of Ephesis is guided by user needs expressed by the project partners.
A first level of partner is the scientific user committee, or CSU. It’s member are INRA researchers and engineers representative of most Plants studied at the INRA, like wheat, maize, grape, pea, trees (fruits and forest), tomatoes.
Phenome, the national center for phenomic, is a key partner to Ephesis. The Ephesis web application will be used as an integrative data portal for Phenome, by data integration and distributed web services. Therefore there will be significant development on Ephesis to fulfill Phenome Needs. Likewise, several species focused projects, like Breedwheat, Amaizing or Rapsodyn, will need to integrate phenotype data in GnpIS, and therefore will request some evolution of GnpIS and the Ephesis application. All those project are directly involved in the developments coordinated by the Ephesis project.
The Trans-national Infrastructure for Plant Genomic Science Europeant project, or Transplant, offers Ephesis a great opportunity. Among other things, this project is setting up a network of phenotype databases, and Ephesis is one of the first node of this network. Ephesis is also highly involved in the construction of standards for Ontologies and data exchange format.
URGI team: Cyril Pommier, Erik Kimmel, Delphine Steinbach, Hadi Quesneville
Help of Unit support team: Erik Kimmel, Mikaël Loaec, Claire Viseux, Aminah Keliet
Duration: 27/04/2010
Coordinator: Cyril Pommier
Creation date: 27 Apr 2010
 eZ Publish
eZ PublishPublication supervisor: A-F. Adam-Blondon
Read Credits & General Terms of Use
Read How to cite








