REPET

- Version: 3.0
- Editor: URGI
- DOWNLOAD
REPET
The REPET package integrates bioinformatics programs in order to tackle biological issues at the genomic scale. It is distributed under
and deposited to the Agence de Protection des Programmes ( APP ) under the Inter Deposit Digital Number FR 001 480007 000 R P 2008 000 31 235.
To download the last REPET package v3.0 . What's new in this release CHANGELOG (19.10 kB) ? Previous releases are available here
For a quick install, REPET and its dependencies are containerized in a free docker image downloadable
NEW! A video tutorial for the usage of the REPET docker image is available here
How to cite REPET package:
The REPET package has evolved with improvements over time. The 3 pipelines of the REPET package are TEdenovo (building TE library), PASTEC (consensus classification) and TEannot (TE annotation).
Here is the full list of publications :
| TEfinder (Blaster, Grouper, Matcher): Quesneville, H., Nouaud, D. & Anxolabéhère, D. Detection of New Transposable Element Families in Drosophila melanogaster and Anopheles gambiae Genomes . J Mol Evol57 (Suppl 1), S50–S59 (2003). https://doi.org/10.1007/s00239-003-0007-2 |
| TEdenovo: Flutre T, Duprat E, Feuillet C, Quesneville H (2011) Considering Transposable Element Diversification in De Novo Annotation Approaches. PLoS ONE 6(1): e16526. https://doi.org/10.1371/journal.pone.0016526 |
| PASTEC (included in TEdenovo): Hoede C, Arnoux S, Moisset M, Chaumier T, Inizan O, Jamilloux V, et al. (2014) PASTEC: An Automatic Transposable Element Classification Tool. PLoS ONE 9(5): e91929. https://doi.org/10.1371/journal.pone.0091929 |
| TEannot: Quesneville H, Bergman CM, Andrieu O, Autard D, Nouaud D, Ashburner M, et al. (2005) Combined Evidence Annotation of Transposable Elements in Genome Sequences. PLoS Comput Biol 1(2): e22. https://doi.org/10.1371/journal.pcbi.0010022 |
| Long join procedure (included in TEannot): Ahmed, I., Sarazin, A., Bowler, C., Colot, V., & Quesneville, H. (2011). Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic acids research, 39(16), 6919-6931. https://doi.org/10.1093/nar/gkr324 |
A methodological article to be able to tackle the TE annotation of large genomes, reduce the resources necessary for the construction of the TE library, improve the consensus library and have metrics on the quality of the annotations (NTE, LTE) is available here:
| V. Jamilloux, J. Daron, F. Choulet and H. Quesneville, "De Novo Annotation of Transposable Elements: Tackling the Fat Genome Issue," in Proceedings of the IEEE, vol. 105, no. 3, pp. 474-481, March 2017, doi: 10.1109/JPROC.2016.2590833. https://ieeexplore.ieee.org/document/7562280 |
Brief description of REPET
Its two main pipelines are dedicated to detecte, annotate and analyse repeats in genomic sequences, specifically designed for transposable elements (TEs).
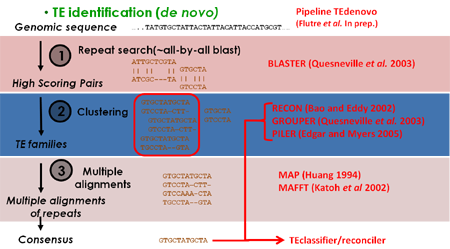
- TEdenovo: this pipeline starts by comparing the genome with itself using BLASTER. Then it clusters matches with GROUPER, RECON and PILER, clustering programs specific for interspersed repeats. For each cluster, it builds a multiple alignment from which a consensus sequence is derived. Finally these consensus are classified according to TE features and redundancy is removed. At the end we obtain a library of classified, non-redundant consensus sequences.
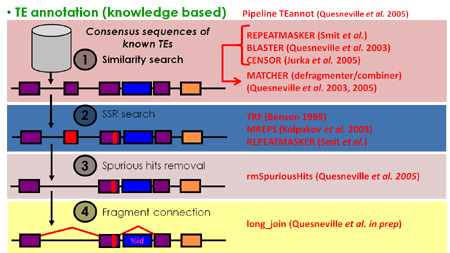
- TEannot: this pipeline mines a genome with a library of TE sequences, for instance the one produced by the TEdenovo pipeline, using BLASTER, RepeatMasker and CENSOR. An empirical statistical filter is applied to discard false-positive matches. Short simple repeats (SSRs) are annotated along the way with TRF, RepeatMasker and MREPS. Then the pipeline chains, with MATCHER via dynamic programming, TE fragments belonging to the same, disrupted copy. A "long join" procedure is subsequently applied to connect distant fragments. Finally annotations are exported into GFF3 and gameXML files.
Repet package also contains PASTEClassifier ( Hoede C. et al. 2014 ) classifies TEs based on Wicker classification ( Wicker T. et al. 2007 )
Prerequisites and installation notes are described in INSTALL
The use of reference banks is optional but much advised to improve your consensus classification.
Currently there is different banks available formatted for a REPET use. Choose your bank according to its content in reference transposable element families on your species to be annotated :
- Here is a link to Repbase :
There is a file specially formatted for REPET ("REPET edition") available with fees.
- Here is a link to REXdb :
The Viridiplantae v3.0 formatted for REPET is available here
- Here is a link to Dfam :
The Dfam3.6 formatted for REPET is available here .
Optional: if you want to search for protein domain by HMM profiles in TE consensus, you need to have hmmpfam (from package hmmer2) or hmmpress and hmmscan (from package hmmer3) and an appropriate bank of HMM profiles ( http://hmmer.janelia.org/ ). It is advisable to use our last version ProfilesBankForREPET_Pfam35.0_GypsyDB.hmm
The old bank version is still available here: ProfilesBankForREPET_Pfam27.0_GypsyDB.hmm.tar.gz bank , specially formatted for REPET.
Help and documentation
To discover and learn how to use REPET pipelines : read TEdenovo and TEannot tutorials, follow practical work .
Development
The development of REPET follows eXtreme programming guidelines since the release 1.3 (in July 2009).
Contact
Reporting bugs or asking for features are much welcome! Please contact us via email at urgi-repet[[@]]inra.fr.
If you want to receive updates, send an email to urgi-repet[[@]]inra.fr with the following information:
- First name
- Last name
- Institution
- Address
- City
- Zip
- Country
- Architecture ( linux-x64 )
- Job scheduling system (SGE or Torque)
Authors and contributors
| Hadi Quesneville | Olivier Inizan |
| Timothée Flutre | Claire Hoede |
| Elodie Duprat | Sandie Arnoux |
| Gaël Faroux | Françoise Alfama |
| Delphine Autard | Jonathan Kreplak |
| Timothée Chaumier | Véronique Jamilloux |
| Marc Bras | Mark Moissette |
| Benoit Bely | Tina Alaeitabar |
| Anna-Sophie Fiston-Lavier | Emmanuelle Permal |
| Erwan Ortie | Emeric Henrion |
| Valentin Marcon | Eric Penneçot |
| Johann Confais | Mariène Wan |
Funding
- The "Modulome" project" funded by

- The
TransPLANT
FP7 european project (EU 7th Framework Programme, contract number 283496)


Access mode(s):
DownloadableKeyword(s):
detection, annotation, transposable element, pipelineCreation date: 14 Oct 2011
 eZ Publish
eZ PublishPublication supervisor: A-F. Adam-Blondon
Read Credits & General Terms of Use
Read How to cite