icons description:
 download subject sequence
download subject sequence
 download subject region corresponding to the HSP
download subject region corresponding to the HSP
 wheat sequences repository
wheat sequences repository
 OakMine PM1N v2.3 Quercus robur annotation database
OakMine PM1N v2.3 Quercus robur annotation database
 Display sequence in GBrowse
Display sequence in GBrowse
 RepetDB datasource or subject concensus card
RepetDB datasource or subject concensus card
 Gramene
Gramene
 URGI -INRAE
URGI -INRAE
 Display Sequence in JBrowse
Display Sequence in JBrowse
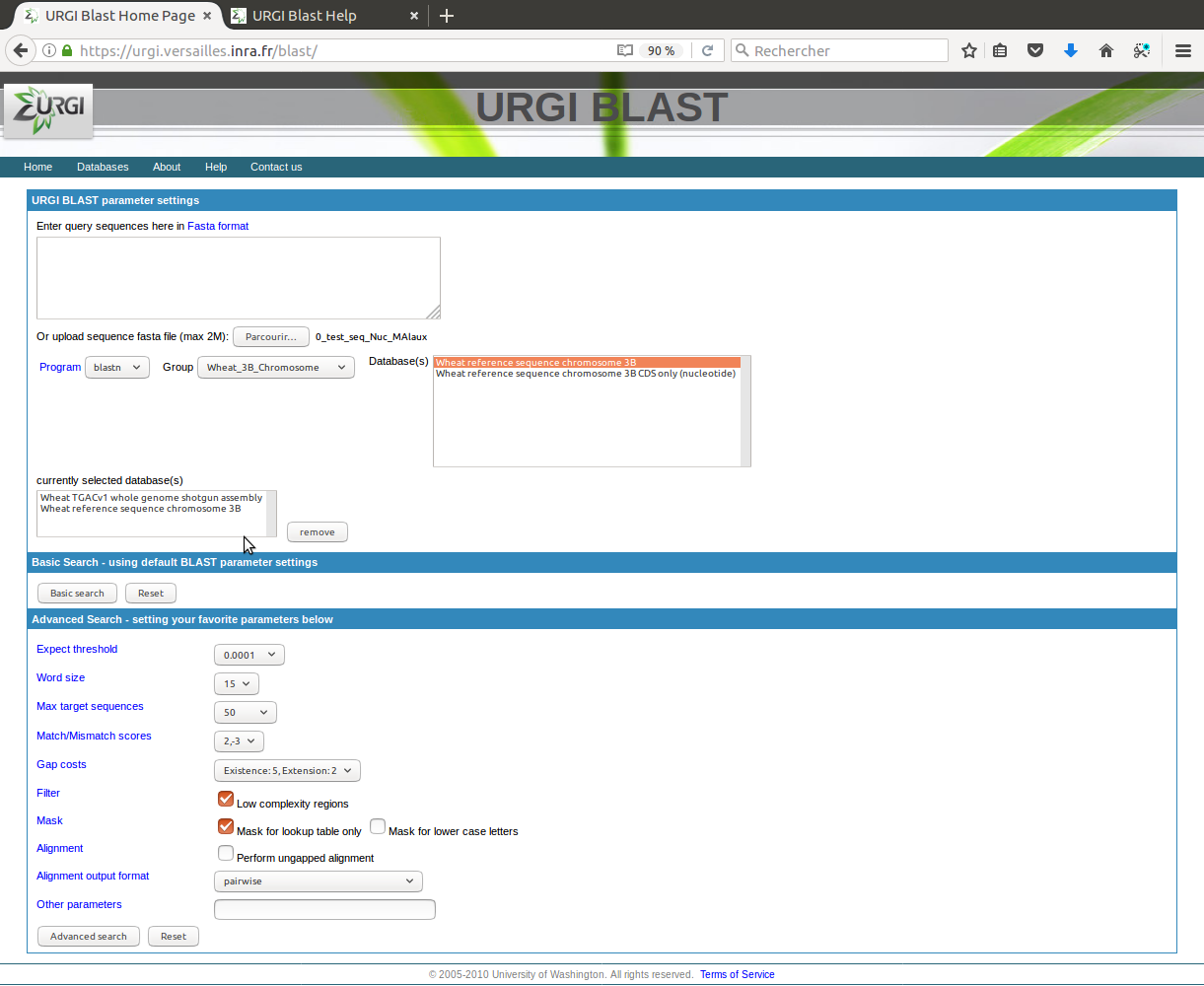
Steps to start a BLAST search:
- Copy and paste multiple query sequences in fasta format to text box, or upload a sequence file in FASTA format.
- Choose a BLAST program, a group of databanks and select one or multiple databases to blast against.
- Select search features to get more specific information (optional).
- Click "Search" button to start search.
Friendly output to easily parse and navigate the BLAST result.
- User can reset different parameters to filter and parse again the BLAST result.
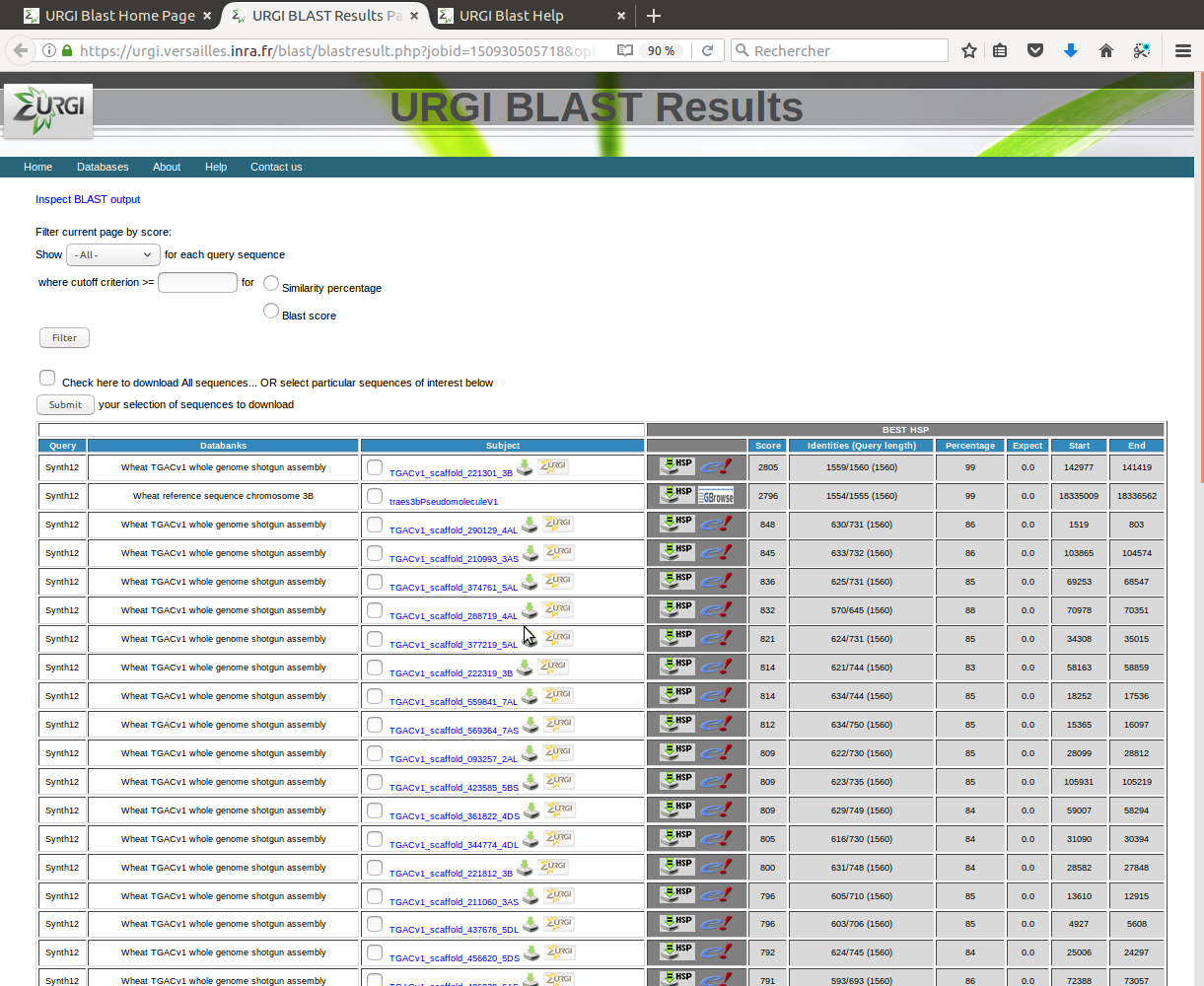
- Subject field could be clickable with link to additional data.
- grey columns are related to data and links of the best hsp.
- Click one of links in Score field will locate the pair-wise alignment between the query sequence and subject sequence in the blast result file.
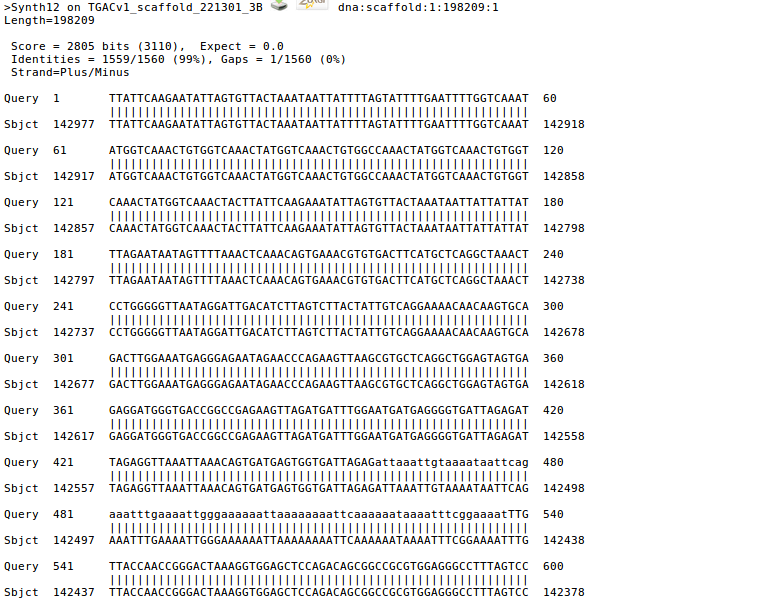
- Click "Inspect BLAST output" link will open the blast result file.
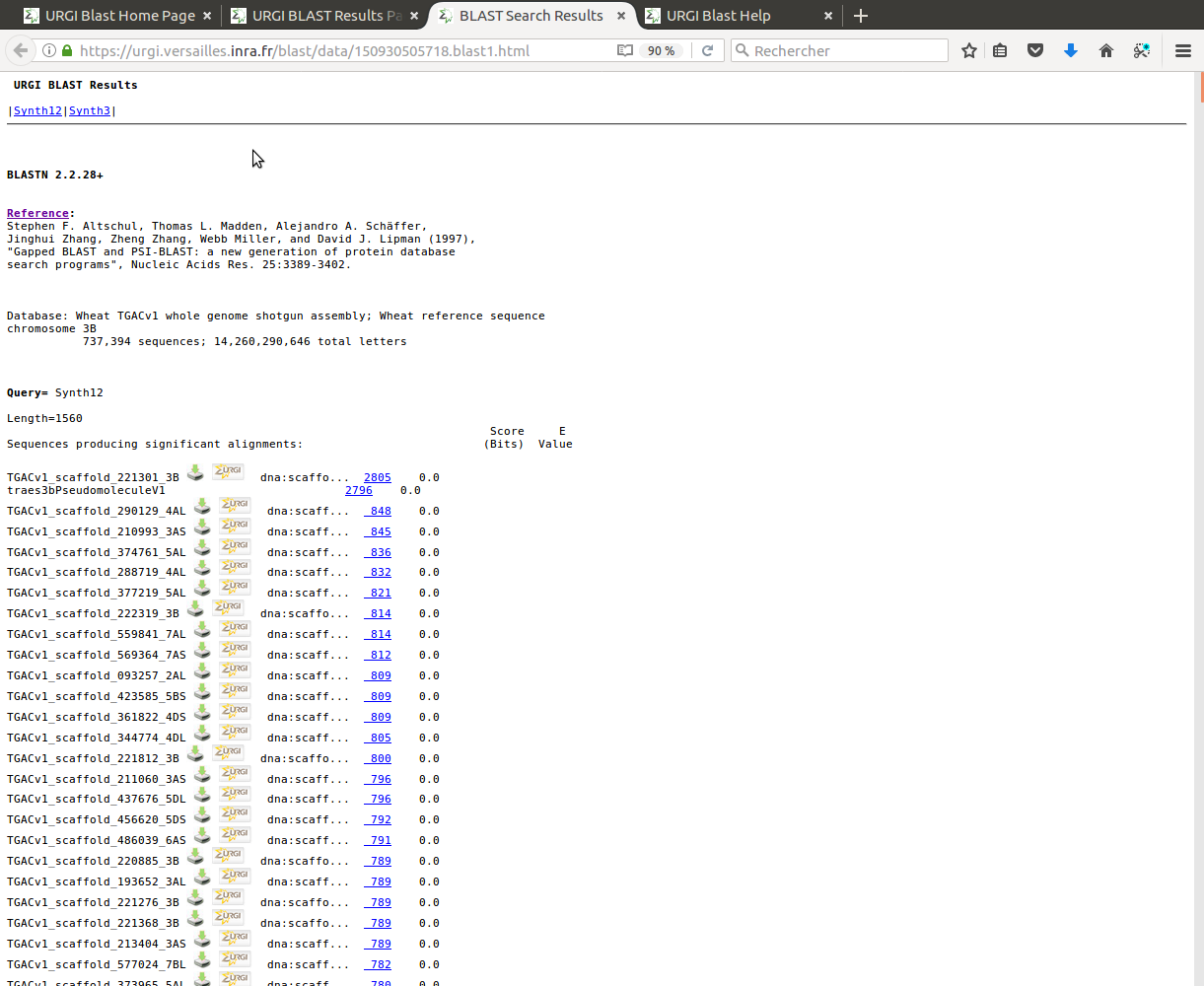
- The tool automatically divides a big output file into several small files to allow users downloading files quickly and easily.
It also provides links to each query sequence blast result for the convenience of analysis of results.
- Check the subject sequences to be downloaded or check to download all sequences and click "Submit" button.
Click "Download" button to download aligned sequences to your local computer.

![]() download subject sequence
download subject sequence![]() download subject region corresponding to the HSP
download subject region corresponding to the HSP![]() OakMine PM1N v2.3 Quercus robur annotation database
OakMine PM1N v2.3 Quercus robur annotation database ![]() RepetDB datasource or subject concensus card
RepetDB datasource or subject concensus card ![]() Display Sequence in JBrowse
Display Sequence in JBrowse 



